Worlds Within Us
Pulled from an unlikely source, ancient microbial DNA represents a new frontier in the study of the past—and modern health
By SAMIR S. PATEL
September/October 2016
The largest ancient DNA laboratory in the United States sits behind a heavy steel door in a plain service hallway at the University of Oklahoma. Inside, researchers find, extract, isolate, and amplify DNA molecules and proteins, producing voluminous mounds of data that can address grand, complex questions about migration, diet, and human health—in the deep past and today. They’re probing the limits of new methodologies. They’re encountering the advantages and pitfalls of interdisciplinary science. And they’re writing the first drafts of a new chapter in archaeological research. But before they can do any of this, they have to ensure that the lab is scrupulously clean.
Next to the door, a red button, when pressed, produces a satisfying thump and turns off powerful UV lights inside. A series of pressure gauges climbs next to it. The lab’s six rooms are kept under positive pressure, double-sealed, and have their own air supply, filtered free of anything larger than 1,000 daltons—the mass of just 1,000 hydrogen atoms. People who enter must take off their shoes, change into scrubs, and, by the time they reach the two innermost rooms, don Tyvek suits, surgical masks, hairnets, and face shields. Those chambers are free of anything extraneous: Only sample vials and scientific equipment are visible. The DNA and proteins that the researchers work with there come from ancient microbes, and keeping the lab free of contamination is a tall order in a world that is positively swimming with their modern counterparts.
“Usually, ancient DNA work is performed in dungeon-like labs located in windowless basements,” says Christina Warinner, anthropologist and codirector of the Laboratories of Molecular Anthropology and Microbiome Research (LMAMR). This lab, however, is fitted with picture windows that face the atrium of the university’s Stephenson Research and Technology Center, so visitors can watch the scientists and students inside process microscopic genetic samples that can be centuries or even millennia old.
The microbes that are the focus of the LMAMR—from both ancient and modern sources, with separate lab facilities for each—come from what is known as the human microbiome, the myriad communities of bacteria (as well as eukaryotes, viruses, and archaea) that reside in and on our bodies. In only the last few years researchers have begun to understand that studying how the microbiome has shifted over thousands of years, particularly at moments of great change in human history, has the potential to reveal some of the ways in which how we eat, live, and move around the world have affected human biology. Any number of questions—medical, archaeological, demographic, evolutionary—that were unframeable just five years ago can now be asked and ultimately answered on scales ranging from molecular to continental.

There are, according to the latest estimates, something like 30 trillion human cells in your body. Alongside them, throughout your gut, in your mouth, on your skin, there are even more—40 trillion, give or take—individual bacteria. Together they make up 2 percent of your body weight, roughly equivalent to the weight of your brain, and carry some 3.3 million genes, to your paltry 22,000. Like every other multicellular organism, we coevolved with them, we incorporated them into our cells, we’re built from them. We may think ourselves individuals, but we’re each a multitude.
The term “microbiome” isn’t yet two decades old but it is already clear that these communities have a profound impact on human health. In addition to critical roles in oral and digestive health, the microbiome has been associated in some way with everything from mood disorders to cardiovascular disease, from autism to rheumatoid arthritis, not to mention countless infectious diseases. The old idea of sickness being caused by individual bugs is now giving way to a much more complex microbial ecosystem model. “On balance it is very clear that the microbiome plays a fundamental biological role,” says Warinner.
The microbiome is a key point of contact between humans and the world around them. It is affected by—and therefore may reflect—changes in how we manipulate our environments and in what we consume. And we change with it. The Neolithic Revolution, for example, saw the rise of agriculture and settlement. The last 150 years have brought the Industrial Revolution, megacities, modern hygiene, processed foods, and antibiotics. They’ve also seen a rise in the incidence of chronic disease: cardiovascular ailments, autoimmune disorders such as asthma and allergies, and metabolic disease such as obesity and diabetes. Experts speculate that changes to the microbiome could be a significant link between lifestyle and health. But examining such connections means knowing how the microbiome has changed—and that means knowing what it was.
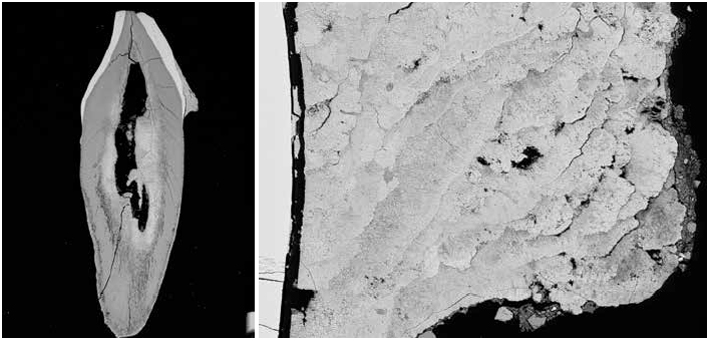
The gut is our most prolific microbial ecosystem, and evidence of ancestral microbiomes—these microbial communities vary by place, time, and culture—though rare, has been found in coprolites, or preserved feces, in particular by LMAMR codirector Cecil M. Lewis, who also studies the modern gut microbiome. But the gut is just one microbial hub in the human body. In your mouth, right now, there are hundreds of species of bacteria, living in nine major niches, including below the gums, on the cheek, and in the saliva. The mouth is the microbial equivalent of a rainforest, teeming with creatures, interspecies warfare, cataclysms. Some of these residents form a film on your teeth, colonies stuck together with DNA, proteins, and polysaccharides. Left unbrushed, this plaque, for reasons that aren’t really known, occasionally fossilizes in your mouth to form tartar, dental calculus. Calculus is tough and almost universally observed clinging to the teeth of adult skeletons discovered at archaeological sites. For many years this material was ignored, discarded, and otherwise overlooked, as were human bones prior to the introduction of modern archaeological practices. “We’ve always thrown stuff away—and that stuff becomes revolutionary,” says Greger Larson, director of the Palaeogenomics & Bio-Archaeology Research Network at the University of Oxford. “If you don’t understand that it exists, then you can’t understand its power.”
When Courtney Hofman, a researcher at the LMAMR who has since joined the full-time faculty there, begins the DNA extraction process, the first thing that registers is the sound. She takes a dental scaler to a 2,000-year-old tooth from Spain and produces the raspy scratching and resonant clinks familiar to anyone who’s had their teeth thoroughly cleaned. After a moment, the tip of the scaler finds purchase and a fragment of calculus pops loose. Hofman does the same with a few more teeth, wiping everything down between each step to limit the possibility of contamination. “We go through a lot of bleach here,” she says.
Each sample goes into a tiny vial to be decontaminated with UV light, and is then crushed, rinsed, and demineralized, leaving behind a gauzy pellet. After a spin in a centrifuge, DNA from 2,000 years ago floats invisibly in each vial. This is the most sensitive part of the process, as the protective calculus is gone. Next, the DNA must be purified and processed for sequencing and analysis.
As a source of ancient biomolecules, calculus differs from coprolites. They contain distinct microbial ecosystems, of course, and preserve very differently. Coprolites are open systems, subject to the elements, contamination, and domination by soil microbes. Calculus, on the other hand, fossilizes while you’re still alive. But it wasn’t clear until very recently—through Warinner’s work and parallel projects in labs in Chile, Australia, and Denmark—that calculus traps or preserves DNA at all. “Dental calculus is really an unlikely hero,” she says.
Warinner, who had excavated the earliest known epidemic mass graves in Mesoamerica early in her career, began to see the potential in calculus in 2007, when she worked at the Smithsonian. A colleague there was Amanda Henry, now at the Max Planck Institute in Leipzig, who searches calculus for trapped plant fragments, such as phytoliths and starch grains. By 2010, Warinner was a postdoctoral researcher studying ancient disease and diet at the Institute of Evolutionary Medicine at the University of Zurich. When she tried to replicate Henry’s methods there, she had trouble counting plant granules. “There were so many bacteria that were getting in the way,” she says, “but I had a kind of aha moment.” Other research groups were exploring the same idea, but at the time nothing had been published, other than a review article from the 1950s that stated that there was no DNA in calculus. She saw an opportunity to test her hypothesis that it could be a source of ancient microbial DNA, but it was a risky, expensive proposition for a postdoc. “People thought it was just a nuts, nuts idea,” Warinner says. “I was sort of doing microbiome research without knowing the word.”
“In German we say überflieger; that means someone who flies higher than the others. I could immediately see her potential,” says Frank Rühli, director of the Institute. But the calculus idea didn’t seem promising. “I was a bit reluctant at the very first moment.” Warinner’s first few attempts in 2010 failed. She then got a more sensitive fluorometer, and it still didn’t appear to be working. “I saw a message that I didn’t even know existed,” she says. “It said ‘Error, DNA too high.’” So she diluted the sample 50 to 100 times. “I started realizing, as I started quantifying, that I had just discovered the richest source of ancient DNA ever described in an archaeological sample.” There was more DNA in her sample than there is in fresh liver tissue.
In the few years before these first experiments, several scientific currents were converging. In 2008, Lewis, who didn’t yet know Warinner, published some of the first reports of the ancient microbiome from coprolites. The same year, the Human Microbiome Project (HMP), a multi-institution effort to catalogue the biota, was established with funding from the National Institutes of Health. This placed the term and its connection to human health in wide view. Microbes had long been seen only as biological villains to be exterminated, but with the launch of the HMP came the recognition that bacteria could be helpful and even essential to multicellular life and its processes. This was a seismic change.
Perhaps most importantly, genetic research itself was undergoing a revolution. Gone were the days of laboriously generating one DNA sequence at a time. The new wave of technology, called next-generation sequencing (NGS), was becoming widely available, and is able to create hundreds of millions of sequences at once. It opened the door for “shotgun metagenomics,” or amplifying and sequencing all the DNA from all the genes from all the organisms in a sample. NGS has made it possible, in a single pass, to describe entire microbial communities or reconstruct the full genome of a microbial species. It increased exponentially the amount of data researchers could gather. According to Warinner, “We were just racing as fast as we could to keep up with all of this.”
NGS, however, was expensive—the necessary reagents alone cost $13,000 for the experiment that Warinner planned to conduct. She was terrified, and thought at the time, “It’s a moon shot. If it works, it’s awesome. If it doesn’t, I’m hosed.”
Dalheim is a small town near Lichtenau, Germany, where in 1989 and 1990 the Westphalian Museum of Archaeology excavated the site of a medieval monastery, parish church, and convent. Remains from the 151 burials they uncovered were stored at the University of Mainz until the school needed the space and planned to incinerate and bury them. Rühli offered to bring the skeletal material to Zurich for research in 2010. “A big challenge when you’re trying to develop a new method is just having material to practice on,” says Warinner. While she was conducting her work on the Dalheim samples, several other groups were operating with the same idea. In 2012, researchers from the University of Chile were the first to publish the basic identification of DNA in archaeological dental calculus. A group led by Alan Cooper of the University of Adelaide was the first to apply NGS to it in 2013. Following these reports, the Dalheim burials offered Warinner an opportunity to produce the first shotgun metagenomic analysis and characterize complete ancient oral microbial communities. The Dalheim study stands now as the most forceful declaration of the possibilities of dental calculus, with orders of magnitude more data than had been reported before. The experiment resulted, Warinner says, in hundreds of millions of sequences. It took three years to analyze it all.
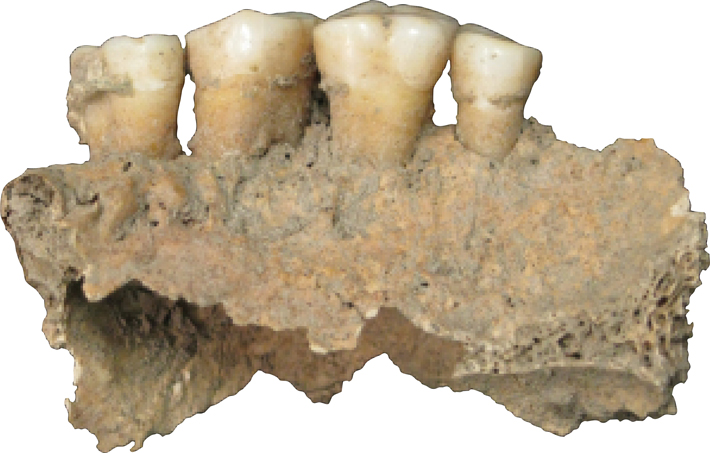 Warinner and her coauthors—32 in total from a broad range of disciplines—catalogued, from the mouths of four medieval individuals, 40 opportunistic pathogens, including species associated with cardiovascular disease, meningitis, and pneumonia, as well as what might be the oral ancestor of modern gonorrhea. They sequenced the entire genome of Tannarella forsythia, a cause of periodontal disease. They saw dietary DNA from pigs, cruciferous vegetables, and bread wheat. They looked for proteins as well, and found ones associated with pathogen virulence, others produced by the human immune system, and beta-lactoglobulin, a durable dairy protein. Among the genes identified, oddly, are ones associated with microbial antibiotic resistance—hundreds of years before the advent of antibiotic drugs. The expert on the subject for the study, Lars Hansen of Aarhus University in Denmark, says that the cellular machinery that creates antibiotic resistance can serve other purposes in cells. The modern phenomenon, he explains, comes from increased availability and expression of these existing mechanisms. “It is the first time [antibiotic resistance sequences] have been found in an ancient human-associated context,” says Hansen. “It shows that these building blocks are basically everywhere.”
Warinner and her coauthors—32 in total from a broad range of disciplines—catalogued, from the mouths of four medieval individuals, 40 opportunistic pathogens, including species associated with cardiovascular disease, meningitis, and pneumonia, as well as what might be the oral ancestor of modern gonorrhea. They sequenced the entire genome of Tannarella forsythia, a cause of periodontal disease. They saw dietary DNA from pigs, cruciferous vegetables, and bread wheat. They looked for proteins as well, and found ones associated with pathogen virulence, others produced by the human immune system, and beta-lactoglobulin, a durable dairy protein. Among the genes identified, oddly, are ones associated with microbial antibiotic resistance—hundreds of years before the advent of antibiotic drugs. The expert on the subject for the study, Lars Hansen of Aarhus University in Denmark, says that the cellular machinery that creates antibiotic resistance can serve other purposes in cells. The modern phenomenon, he explains, comes from increased availability and expression of these existing mechanisms. “It is the first time [antibiotic resistance sequences] have been found in an ancient human-associated context,” says Hansen. “It shows that these building blocks are basically everywhere.”
The study has spun off in dozens of directions. For example, the LMAMR is using the presence of the milk protein to study the origins of dairying practices all over the world. Another of their studies announced the reconstruction of a full human mitochondrial genome from calculus alone. This affirms calculus as a possible alternative source of human DNA in cases, such as with Native American remains, where it is not permissible to sample bone. Archaeologist Mark Aldenderfer from the University of California, Merced, along with Warinner and her team, is studying the genetic adaptations that allow people to live and thrive at high altitudes in Nepal and Peru. The LMAMR is also collaborating with archaeologists who work in the Caribbean to see how well calculus preserves DNA in climates generally unfriendly to ancient biomolecules, and to study migration and colonialism alongside archaeological and linguistic evidence. And in the study that Hofman is working on, some 20,000 years’ worth of samples from a site in Spain are being examined to look for changes in pathogens, proteins, and microbial community structure over time in a single place. The origins and evolution of specific diseases can be examined as well—typhoid, tuberculosis, plague, syphilis. “We don’t know what the limits are yet,” says Warinner.
Another step in this research concerns the study of proteins, or proteomics. Proteins, which are also found in calculus, may persist much longer than DNA and reflect the actual expression of genes. According to Lewis, “That’s going to change everything.” One thing that proteomics may be able to show is direct interaction between microbes and the human immune system. Matthew Collins, an LMAMR collaborator and specialist in ancient proteins with the Universities of York and Copenhagen, compares proteins in calculus to the port of an ancient city destroyed suddenly and preserved in situ. The calculus holds evidence, in proteins, of ongoing battles between “migrant workers” and the “local police force.” “The body fights against infection and you can see the bugs fighting back,” he explains. “You’ve got this whole dynamic system preserved there.” The work will require innovation and refinement in extraction and analysis, but the potential is vast.
Research is going forward, but it remains in its early stages, and will have notable pitfalls and hurdles to overcome. The LMAMR, working with Kirsten Ziesemer of Leiden University in the Netherlands, has already identified one of these problems. Apparently a gene segment called 16s, commonly used to identify species from their DNA, can’t be relied on for conventional genetic analysis of ancient microbes because of the way it breaks down over time. The array of processes and methods is complicated and has yet to be standardized. “One of the problems has been, in microbiome research in general, that different methods of extraction produce different results, different computational pipelines produce different results, different sequencing platforms produce different results, different primer sets produce different results,” says Camilla Speller, another LMAMR collaborator from the University of York. There’s a lot of troubleshooting before innovation can become practice. “This is a problem,” Lewis says, “of being on the frontier.”
At the LMAMR, after the calculus has been demineralized, the scale of the work goes from tiny to microscopic to molecular. The DNA floating in the solution has been broken down by time into fragments, from just a few base pairs up to maybe 100 in length. Conveniently, next-generation sequencers are designed to work on DNA fragments of about that length, around 50 to 100 base pairs. The samples are treated with a series of enzymes, buffers, and primers, and attached to segments of synthetic DNA. These manmade genetic fragments, which can later be excised from the data, are used to fill in gaps, tag each sample with a unique genetic barcode, and make the samples machine-readable. This process is called “building a library.” The library is then “read” at a genomics facility, such as the Yale Center for Genome Analysis, and what comes back is a stream—a tsunami, really—of data.
The quantity is intimidating. “I wouldn’t say it’s a wall, but I’d say it’s a very steep mountain,” says Warinner. According to LMAMR codirector Krithi Sankaranarayanan, who is a microbial ecologist adept at data analysis and interpretation, the next steps also include confirming, on the basis of damage patterns, that the DNA is indeed old. The reads are then assembled and modeled and interpreted to create longer and longer sequences: partial genes, full genes, and even complete genomes. Then the researchers must try to manage and corral these massive data sets to make them comprehensible. They pool them, build frequency tables, parse species into different bacterial groups. They look for questions and structures and layers that bring order. Modern computing power is staggering, but analysis still takes days or weeks.
If the whole thing sounds Borgesian, that’s because it is. Jorge Luis Borges imagined the universe, in his 1941 short story “The Library of Babel,” as an infinite library containing every possible combination of the alphabet—an expanse of gibberish concealing magical insights. The librarians there are wandering mendicants driven to superstition, madness, and worse. In today’s research, even a single sample of calculus creates tomes upon tomes, composed of patterns of four letters representing the nucleobases that make up DNA: A, G, C, and T. There is a seemingly limitless, global, paralyzing abundance of potential samples in museums, archaeological collections, and unexcavated sites. Complicating matters, the databases against which these DNA patterns are checked are imperfect, skewed toward specific species that have been studied because of their potential impact on human health or agriculture. There’s also DNA “dark matter,” or sequences that don’t match up with anything that has been described or characterized—undiscovered biology. Interpretation requires knowledge of these flaws, computational mastery, a global perspective, and savage, callous skepticism.
Taking data sets so large that they can only be contemplated through summary statistics or pure abstraction and working them into the discipline of archaeology is a challenge. But this new methodology certainly isn’t the first time that archaeologists have had to come to grips with scientific innovation. Radiocarbon dating, isotopic signatures, and remote sensing have all become regular analytical tools. Similarly, geneticists have to begin to acknowledge and understand the language and questions of archaeologists.
“There have been revolutions in archaeology before, and archaeologists have been able to adapt to them,” says Hannes Schroeder, who studies ancient DNA and the Caribbean at the Natural History Museum of Denmark, and is also working with LMAMR. “But none of these other fields are as demanding in analytics, techniques, and working with the data as what is happening now with ancient DNA.” For York’s Speller, it means that archaeology is growing in a new direction, one that includes computer science and medical bioscience.
Warinner and Lewis at the LMAMR, and a number of other researchers looking at the ancestral microbiome, have backgrounds and experience in archaeology and anthropology. “The goal is to find common questions we’re really interested and invested in,” says Warinner. “The challenge of a scientist is to chart a path through these questions so you don’t get totally overwhelmed by them. It is such enormous territory.”
At the moment, the areas most suited to examination through the lens of ancestral microbiomes are big ones—the peopling of continents, agriculture, migration, exploration, colonialism, industry, globalism. Researchers want to know what these transitions mean for the shape of the modern world—specifically, who we were and who we are, physically, genetically, microbially. We can now investigate patterns, identify specific bacteria or entire microbial communities that have been lost, and attempt to understand what they did for us after tens of thousands of years of coevolution. We might also ask whether we can get them back, or if we even want to. “Now that we know so much about the microbiome, it’s very hard to think of ourselves solely from the perspective of our genome,” says Warinner, “because if you just have genomic information you’re missing such a huge part of the biology that it’s almost irresponsible not to consider it.”
In the late seventeenth century, Dutch draper and scientist Antonie van Leeuwenhoek looked through one of his pioneering microscopes at a sample from inside his own mouth. He found it crawling with what he called “animalcules.” Science perhaps still has moments of revelation, but more progress comes from patience, technological refinement, some new algorithm or process, and ways to think around dead ends and blind alleys. Innovations developed this way build upon one another to a point at which we can see a gene associated with antibiotic resistance in the mouth of a medieval German nun, and this helps us understand something about cellular machinery—machinery that happens to be at the root of a modern healthcare crisis. “I feel,” says Warinner, “like we’re standing at the beginning of a new branch of archaeology.”
Samir S. Patel is deputy editor at ARCHAEOLOGY.



















































































